TACGenomics is a genomic service company based in California USA, providing comprehensive solution to the problem of handling data generated by NGS devices.

- 1 Quality Control of the Data
- 1.1 Quality Control of Raw Fastq Data
- 1.2 Quality Control of Mapping Statistics Data
- 1.3 Quality Control of Coverage Data
- 1.4 Quality Control of Sample PCA Data
- 2 Analyzed Result
- 2.1 Normalized Gene Expression Data
- 2.2 Differential Gene Expression Data
- 2.3 Analyzed GO and Pathway Data
- 2.3.1 GO Enrichment Analysis
- 2.3.2 KEGG Pathway Enrichment Analysis
- 2.3.3 Reactome Pathway Enrichment Analysis
- 3 Methods Description
- 4 Reference
RNAseq Data Analysis Report
1 Quality Control of the Data
1.1 Quality Control of Raw Fastq Data
QC evaluation of fastq data
| Filename | Total_Sequences | Sequence_Length | GC_Percentage | QC_Detail |
|---|---|---|---|---|
| Cancer1_R1.fastq.gz | 42388539 | 50-150 | 47 | See detail |
| Cancer1_R2.fastq.gz | 42388539 | 50-150 | 47 | See detail |
| Cancer2_R1.fastq.gz | 42641519 | 50-150 | 48 | See detail |
| Cancer2_R2.fastq.gz | 42641519 | 50-150 | 49 | See detail |
| Cancer3_R1.fastq.gz | 41688137 | 50-150 | 49 | See detail |
| Cancer3_R2.fastq.gz | 41688137 | 50-150 | 50 | See detail |
| Normal1_R1.fastq.gz | 41361293 | 50-150 | 47 | See detail |
| Normal1_R2.fastq.gz | 41361293 | 50-150 | 47 | See detail |
| Normal2_R1.fastq.gz | 37607001 | 50-150 | 49 | See detail |
| Normal2_R2.fastq.gz | 37607001 | 50-150 | 49 | See detail |
1.2 Quality Control of Mapping Statistics Data
QC evaluation of mapping data
| Sample | Total_Reads | Mapped_Reads | Ribsome_Reads | Coding_Reads | UTR_Reads | Intronic_Reads | Intergenic_Reads |
|---|---|---|---|---|---|---|---|
| Normal1 | 41361293 | 38781291 (93.76%) | 25022 (0.07%) | 22138959 (57.58%) | 8466157 (22.02%) | 5039534 (13.11%) | 2780231 (7.23%) |
| Cancer1 | 42388539 | 39161503 (92.38%) | 12164 (0.03%) | 20214771 (52.14%) | 9806043 (25.29%) | 5109707 (13.18%) | 3630304 (9.36%) |
| Normal2 | 37607001 | 34450687 (91.60%) | 19065 (0.06%) | 18123292 (52.84%) | 6920256 (20.18%) | 4284096 (12.49%) | 4953391 (14.44%) |
| Cancer2 | 42641519 | 38221687 (89.63%) | 12040 (0.03%) | 17632951 (46.28%) | 9837193 (25.82%) | 5402288 (14.18%) | 5214043 (13.69%) |
| Normal3 | 36343292 | 32878384 (90.46%) | 6094 (0.02%) | 18289264 (55.85%) | 7133801 (21.79%) | 3162628 (9.66%) | 4153693 (12.68%) |
| Cancer3 | 41688137 | 38190233 (91.60%) | 13258 (0.03%) | 20601686 (54.19%) | 10154672 (26.71%) | 3348380 (8.81%) | 3897945 (10.25%) |
1.3 Quality Control of Coverage Data
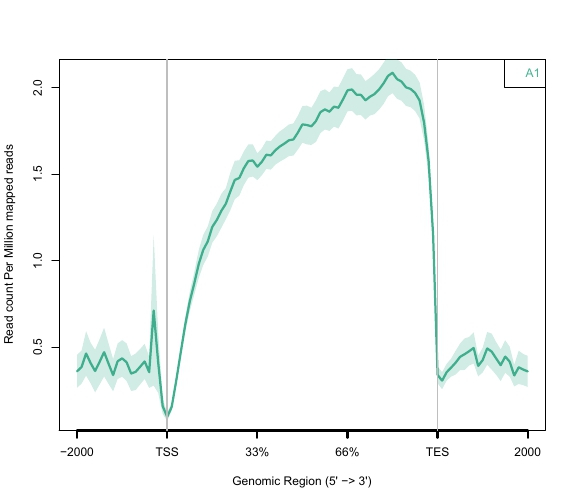
Cancer1 Gene Body Coverage Plot
Cancer2 Gene Body Coverage Plot
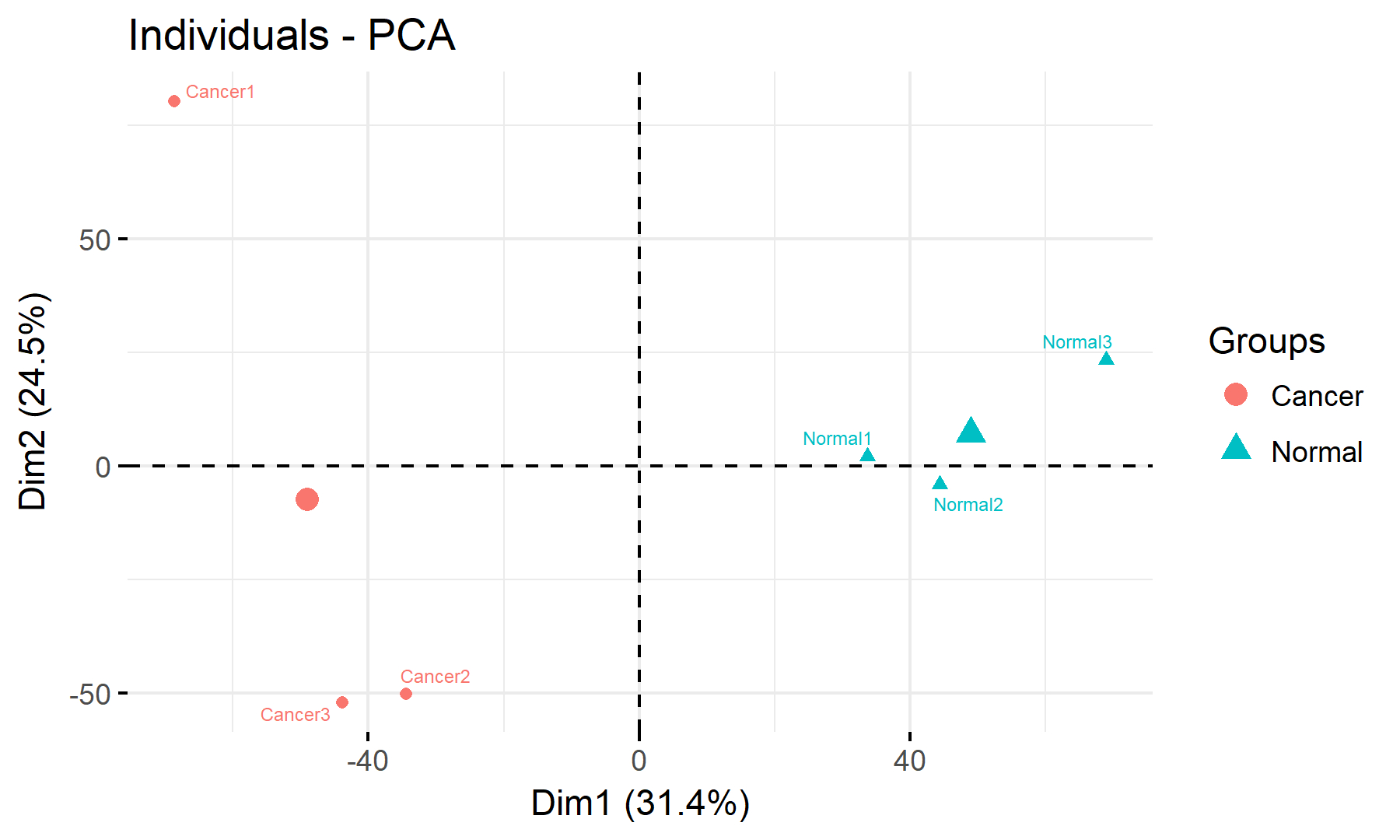
1.4 Quality Control of Sample PCA Data
PCA Plot
2 Analyzed Result
2.1 Normalized Gene Expression Data
TMM normalized gene expression data
| Gene_symbol | NAME | Cancer1 | Cancer2 | Cancer3 | Normal1 | Normal2 | Normal3 |
|---|---|---|---|---|---|---|---|
| A1BG | alpha-1-B glycoprotein | 131.205 | 61.253 | 852.089 | 856.024 | 968.001 | 892.122 |
| A1BG-AS1 | A1BG antisense RNA 1 | 1.995 | 1.124 | 3.404 | 1.670 | 2.528 | 3.062 |
| A1CF | APOBEC1 complementation factor | 777.165 | 228.392 | 702.826 | 759.908 | 755.408 | 1278.096 |
| A2M | alpha-2-macroglobulin | 1082.644 | 635.069 | 3665.401 | 8478.250 | 9713.626 | 9161.569 |
| A2M-AS1 | A2M antisense RNA 1 (head to head) | 1.848 | 1.760 | 2.618 | 3.898 | 8.028 | 7.931 |
| A2ML1 | alpha-2-macroglobulin-like 1 | 0.000 | 0.073 | 0.029 | 0.000 | 0.089 | 0.100 |
| A2MP1 | alpha-2-macroglobulin pseudogene 1 | 0.029 | 0.147 | 0.058 | 0.139 | 0.310 | 0.000 |
| A3GALT2 | alpha 1,3-galactosyltransferase 2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.089 | 0.050 |
| A4GALT | alpha 1,4-galactosyltransferase | 0.528 | 3.569 | 1.542 | 4.454 | 2.262 | 2.460 |
| A4GNT | alpha-1,4-N-acetylglucosaminyltransferase | 0.000 | 1.198 | 0.407 | 0.278 | 0.044 | 0.151 |
2.2 Differential Gene Expression Data
CancervsNormal differential gene expression data
| Gene_symbol | NAME | Foldchange | PValue | FDR | Cancer1 | Cancer2 | Cancer3 | Normal1 | Normal2 | Normal3 |
|---|---|---|---|---|---|---|---|---|---|---|
| A2M | alpha-2-macroglobulin | -5.185 | 0.007 | 0.068 | 1075.741 | 640.041 | 3612.968 | 8485.054 | 9782.632 | 9363.327 |
| A2M-AS1 | A2M antisense RNA 1 (head to head) | -3.239 | 0.014 | 0.111 | 1.837 | 1.774 | 2.581 | 3.901 | 8.085 | 8.106 |
| AACS | acetoacetyl-CoA synthetase | 3.165 | 0.005 | 0.053 | 19.998 | 14.091 | 14.022 | 7.592 | 3.931 | 3.643 |
| AADAC | arylacetamide deacetylase | -2.980 | 0.004 | 0.042 | 73.055 | 94.644 | 166.397 | 373.973 | 271.236 | 350.353 |
| AADACP1 | arylacetamide deacetylase pseudogene 1 | -2.984 | 0.038 | 0.213 | 2.216 | 4.828 | 1.348 | 3.587 | 7.058 | 14.468 |
| AADAT | aminoadipate aminotransferase | -9.155 | 0.000 | 0.000 | 6.384 | 8.967 | 2.437 | 30.508 | 45.295 | 87.268 |
| AASS | aminoadipate-semialdehyde synthase | -5.144 | 0.001 | 0.020 | 112.469 | 33.970 | 7.599 | 305.886 | 202.846 | 283.607 |
| AATK | apoptosis-associated tyrosine kinase | -3.563 | 0.012 | 0.100 | 0.787 | 0.961 | 0.602 | 1.881 | 4.333 | 2.206 |
| ABAT | 4-aminobutyrate aminotransferase | -6.354 | 0.000 | 0.004 | 286.623 | 58.259 | 79.026 | 908.673 | 704.669 | 1080.044 |
| ABCA8 | ATP-binding cassette, sub-family A (ABC1), member 8 | -8.943 | 0.000 | 0.000 | 22.243 | 14.707 | 4.359 | 120.188 | 85.588 | 163.761 |
CancervsNormal Vocalno Plot
CancervsNormal Heatmap Plot
2.3 Analyzed GO and Pathway Data
2.3.1 GO Enrichment Analysis
CancervsNormal GO enrichment analysis data
| GO_ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | Count |
|---|---|---|---|---|---|---|---|
| GO:0016054 | organic acid catabolic process | 99/1743 | 213/16536 | 0 | 0 | 0 | 99 |
| GO:0046395 | carboxylic acid catabolic process | 99/1743 | 213/16536 | 0 | 0 | 0 | 99 |
| GO:0044282 | small molecule catabolic process | 114/1743 | 337/16536 | 0 | 0 | 0 | 114 |
| GO:1901565 | organonitrogen compound catabolic process | 106/1743 | 347/16536 | 0 | 0 | 0 | 106 |
| GO:0071103 | DNA conformation change | 84/1743 | 251/16536 | 0 | 0 | 0 | 84 |
| GO:0008202 | steroid metabolic process | 91/1743 | 287/16536 | 0 | 0 | 0 | 91 |
| GO:0006334 | nucleosome assembly | 56/1743 | 139/16536 | 0 | 0 | 0 | 56 |
| GO:0065004 | protein-DNA complex assembly | 67/1743 | 190/16536 | 0 | 0 | 0 | 67 |
| GO:0071466 | cellular response to xenobiotic stimulus | 63/1743 | 175/16536 | 0 | 0 | 0 | 63 |
| GO:0071824 | protein-DNA complex subunit organization | 71/1743 | 217/16536 | 0 | 0 | 0 | 71 |
- ID: the ID of the GO Term
- Description: the description of the GO Term
- GeneRatio: the denominator is the number of genes within that list which are annotated to the GO of interest and the numerator is the size of the list of genes of interest
- BgRatio: the denominator is the number of genes that are annotated to the GO of interest and the numerator is the total number of genes in the background distribution
- pvalue: P-value in hypergenometric test
- Count: The number of differential genes in this GO Term
GO enrichment bar plot shows most top 50 significant GO Terms.
CancervsNormal GO enrichment Plot
2.3.2 KEGG Pathway Enrichment Analysis
CancervsNormal KEGG Pathway enrichment analysis data
| PATH_ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | Count |
|---|---|---|---|---|---|---|---|
| hsa05322 | Systemic lupus erythematosus | 57/951 | 133/7469 | 0 | 0 | 0 | 57 |
| hsa00280 | Valine, leucine and isoleucine degradation | 30/951 | 48/7469 | 0 | 0 | 0 | 30 |
| hsa04610 | Complement and coagulation cascades | 36/951 | 79/7469 | 0 | 0 | 0 | 36 |
| hsa00071 | Fatty acid degradation | 25/951 | 44/7469 | 0 | 0 | 0 | 25 |
| hsa04110 | Cell cycle | 45/951 | 124/7469 | 0 | 0 | 0 | 45 |
| hsa05034 | Alcoholism | 57/951 | 180/7469 | 0 | 0 | 0 | 57 |
| hsa00260 | Glycine, serine and threonine metabolism | 22/951 | 40/7469 | 0 | 0 | 0 | 22 |
| hsa00380 | Tryptophan metabolism | 22/951 | 40/7469 | 0 | 0 | 0 | 22 |
| hsa01200 | Carbon metabolism | 41/951 | 116/7469 | 0 | 0 | 0 | 41 |
| hsa01212 | Fatty acid metabolism | 22/951 | 48/7469 | 0 | 0 | 0 | 22 |
CancervsNormal KEGG Pathway enrichment analysis data
CancervsNormal KEGG Pathway enrichment analysis data
CancervsNormal KEGG Pathway Bubble Plot
2.3.3 Reactome Pathway Enrichment Analysis
CancervsNormal Reactome Pathway enrichment analysis data
| PATH_ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | Count |
|---|---|---|---|---|---|---|---|
| hsa05322 | Systemic lupus erythematosus | 57/951 | 133/7469 | 0 | 0 | 0 | 57 |
| hsa00280 | Valine, leucine and isoleucine degradation | 30/951 | 48/7469 | 0 | 0 | 0 | 30 |
| hsa04610 | Complement and coagulation cascades | 36/951 | 79/7469 | 0 | 0 | 0 | 36 |
| hsa00071 | Fatty acid degradation | 25/951 | 44/7469 | 0 | 0 | 0 | 25 |
| hsa04110 | Cell cycle | 45/951 | 124/7469 | 0 | 0 | 0 | 45 |
| hsa05034 | Alcoholism | 57/951 | 180/7469 | 0 | 0 | 0 | 57 |
| hsa00260 | Glycine, serine and threonine metabolism | 22/951 | 40/7469 | 0 | 0 | 0 | 22 |
| hsa00380 | Tryptophan metabolism | 22/951 | 40/7469 | 0 | 0 | 0 | 22 |
| hsa01200 | Carbon metabolism | 41/951 | 116/7469 | 0 | 0 | 0 | 41 |
| hsa01212 | Fatty acid metabolism | 22/951 | 48/7469 | 0 | 0 | 0 | 22 |
Reactome enrichment plots show most top 15 significant Reactome pathway.
CancervsNormal Reactome Pathway Bar Plot
CancervsNormal Reactome Pathway Bubble Plot
3 Methods Description
4 Reference
[1] Langmead B et al., Fast gapped-read alignment with Bowtie 2, Nat Methods 9:357-9 (2012)
[2] Li B et al., RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome, BMC Bioinformatics 12:1-16 (2011)
[3] Robinson MD et al, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics 26:139-140 (2010)
[4] Matthew DY et al., Gene ontology analysis for RNA-seq: accounting for selection bias, Genome Biology 11:R14 (2010)
[5] Chen X et al., KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases, Nucleic Acids Res 39:316-322 (2011)
Contact us
- 1-(424)-361-5063
- service@tacgenomics.com
- 425 Broadway, Santa Monica, CA 90401 USA
© 2023 TAC Genomics. All Rights Reserved. Developed By Digital Guider.